We haven’t published a Castor Research Award nomination in a while and this is mostly to keep the suspense going 🙂 Since we have received many submissions, we will be posting more frequently in the coming period so that by the end of the year we are ready for our grand finale!
If last time we nominated a surgical research project, this time we are zooming in into epigenetics – a quite intriguing topic, especially when looking at its role in immunity! How does DNA methylation influence the innate immune response? Is the microbiome altered in patients with pneumonia? Does the microbiome influence DNA methylation in immune cells? The ELDER-BIOME study aims to answer these questions and further elucidate the intricate symbiosis we nurture with bacteria.

![]()
ELDER-BIOME
The Effect of Leukocyte Dna mEthylation and micRoBIOME diversity on host defense mechanisms during community-acquired pneumonia
Who are we?
It is our pleasure to officially submit a project statement for the Castor Research Award. We are Xanthe Brands and Bas Haak, two PhD candidates that aim to investigate the role of leukocyte DNA methylation and the intestinal microbiota on the pathogenesis of community-acquired pneumonia.
This project is supervised by prof. dr. Tom van der Poll, prof dr. Joost Wiersinga and dr. Brendon Scicluna of the Center of Experimental and Molecular Medicine department of the Academic Medical Center.

Our team consists of the following members (from left to right): Stijn Klarenbeek (lab technician), Tom van der Poll (principal investigator), Tjitske van Engelen (PhD candidate), Joost Wiersinga (co-investigator), Bas Haak (main applicant, PhD candidate), Xanthe Brands (main applicant, PhD candidate), Niki Achterkamp (data analist), Mentxu Sanchez Serra (lab technician), Idysha Perez Sanchez (intern), Natasja Otto (PhD candidate). Not depicted: Rosan van der Lee (lab technician), Brendon Scicluna (co-investigator), Daan Faber (principal investigator BovenIJ hospital)
Background
Community-acquired pneumonia (CAP) represents a major health care problem and mortality and morbidity associated with severe pneumonia remain considerable, despite state of the art care. (1) Therefore, there is an urgent need to expand our knowledge on the pathogenesis of pneumonia.
Epigenetics and DNA methylation
All cells within an organism contain the same DNA sequence (genetic code). To ensure the correct activation or deactivation of cellular transcriptional programs, specific regions of the genome require varying degrees of accessibility to RNA polymerases and transcription factors. In a stimulus-specific mode chromatin juggles between an accessible state (euchromatin) to a closed/packed state (constitutive/facultative heterochromatin). These heritable modifications to the chromatin landscape from one cell generation to the next are termed epigenetic modifications. Thus, epigenetics encompasses the study of mitotically and/or meiotically heritable modifications to gene function that are not explained by changes in the DNA sequence. (2)
Various factors can influence DNA methylation status at gene loci, including ageing, nutrition, air pollution and tobacco smoke (3, 4). In addition, microbes can affect DNA methylation, thereby causing infection-induced host gene reprogramming. It has been shown that pathogens can modify DNA methylation patterns in several cell types, and these changes have functional consequences for the capacity of cells to activate essential innate immune pathways (5). Further evidence supports the concept that alterations in DNA methylation patterns have functional consequences for host defense against infection, at least in part by modifying the capacity to activate the master regulator of innate immunity, nuclear factor (NF)kB. (6)
The gut microbiota primes systemic immunity
Spectacular advances have been made in our understanding of the microbiota in health and disease. The gut microbiota can be seen as an exteriorized organ that exerts numerous beneficial functions in the development of the immune system and local support of mucosal immunity (7, 8). Furthermore, the microbiota protects against epithelial cell injury and pathogen colonization (7, 8).
Our group has recently shown that the gut microbiota play a protective role during S. pneumoniae pneumonia as evidenced by the increased bacterial dissemination, inflammation, organ failure and mortality in microbiota-depleted mice compared with controls. Fecal Microbiota Transplantation (FMT) to gut microbiota-depleted mice led to a normalization of pulmonary bacterial counts and TNF-α and IL-10 levels 6 hours after pneumococcal infection (9).
These findings are in accordance with other studies that showed that microbiota-derived products translocate to the circulation and remotely primes bone marrow neutrophils who then show an increased capacity to kill microorganisms (10). A schematic overview of microbiota-derived systemic immunity is depicted in figure 1.
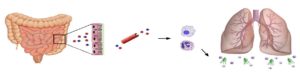
While the role of altered DNA methylation in cancer has been widely studied, knowledge of its impact on antibacterial defense in humans is highly limited. In addition, how the gut microbiota contributes to host defense against bacterial pneumonia has not yet been elucidated in humans. This study aims to shed light on these processes in the contact of community-acquired pneumonia.
What are our objectives?
This study aims to explore a completely novel research area linking the extent of DNA methylation in blood leukocyte (monocytes and neutrophils) and function of gut microbiota on the influence of innate immune responses to and host defense against CAP. Our objectives are as follows:
1. To obtain insight in the role of altered DNA methylation in blood leukocytes (monocytes and neutrophils) in innate immune responses and host defense in patients with CAP.
2. To determine the composition and function of the gut and respiratory microbiota in patients with CAP.
3. To assess the influence of the gut microbiota on leukocyte DNA methylation in patients with CAP.
Study outline and methods
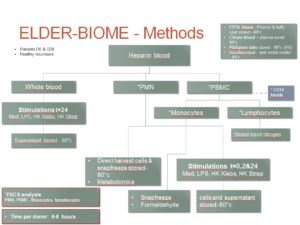
We conduct a multicenter observational study among patients with CAP at the Emergency Department, Intensive Care unit and Internal Medicine Ward of the Academic Medical Center Amsterdam and BovenIJ ziekenhuis. We aim to include a total of 231 CAP patients and 115 healthy subjects above 18 years of age. A maximum of 125 ml of blood will be drawn to analyze DNA methylation patterns of purified monocytes and neutrophils, which will be analyzed in connection with DNA methyltransferase and ten eleven translocation (TET) activity, RNA and DNA gene expression and a selection of standard innate immune function, as well as coagulation tests.
Moreover, isolated monocytes and neutrophils from admission samples will be stimulated in vitro with lipopolysaccharide (LPS), Streptococcus pneumoniae and Klebsiella pneumoniae. In addition, rectal and nasopharyngeal swabs will be obtained to investigate the role of the gut and respiratory microbiota composition and function. Patient material will be obtained upon inclusion and on day 28, when patients will be seen in the outpatient clinic for follow up.
How do we use Castor?
Castor EDC will play an invaluable role in collecting clinical metadata of all study participants. The main parameters of this study will be the alterations in leukocyte DNA methylation and the composition and function of the gut and respiratory microbiota in patients with CAP. These markers will subsequently be associated with several clinical parameters, which are documented in Castor, including, but not limited to:
• Previous medical history of study participants
• Chronic and clincial medication data
• Dietary habits of participants
• Vaccination status
• Date of hospital admission and discharge, transfer or death
• Laboratory findings during admission
• Microbiology results during CAP admission
• Bristol Stool Scale
• Antibiotics, antivirals, and antifungal exposure up to 90 days prior to hospital admission
• Date of high care unit/intensive care unit admission and discharge, transfer or death, if applicable
• Pneumonia disease severity scores, such as the PSI score, qSOFA score and the CURB-65 score
References
1. Organization. WH (2014) http://www.who.int/mediacentre/factsheets/fs310/en.
2. Shaw AC, Goldstein DR, & Montgomery RR (2013) Nat Rev Immunol 13(12):875-887.
3. Scourzic L, Mouly E, & Bernard OA (2015) Genome medicine 7(1):9.
4. Tahiliani M, et al. (2009) Science 324(5929):930-935.
5. Takahashi K (2014) Cell Mol Life Sci 71(6):1045-1054.
6. O’Gorman A, Colleran A, Ryan A, Mann J, & Egan LJ (2010) American journal of physiology 299(1):G96-G105.
7. Schuijt TJ, van der Poll T, de Vos WM, Wiersinga WJ. Trends in microbiology 2013;21:221-9.
8. Clemente JC, et al. Cell 2012; 148:1258-1270
9. Clarke TB, et al. Nat Med 2010; 16:228-231
10. Schuijt T, Lankelma JM, Gut. 2016 Apr;65(4):575-83.
Stay tuned for more nominations! We wish all nominees good luck and encourage all researchers to brag about their projects and get the chance to win €3000!


